
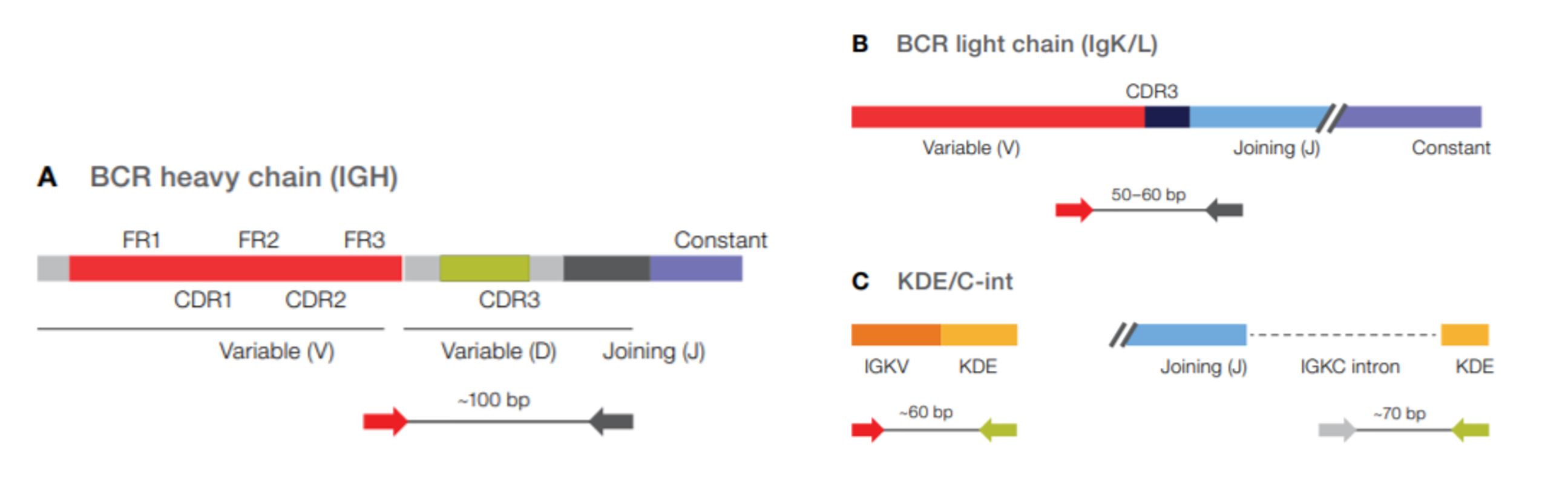
Recent estimates suggest that naïve T cell clone can be as small as ~ 5 cells, which constitutes a negligible fraction of ~ 3 × 10 11 T cells in the human body. Moreover, the frequency of antigen-specific T cells in the naive pool determines both the magnitude and the kinetics of the cognate immune response. Using this approach, it has been shown that the absolute numbers of specific T cells in the pre-immune repertoire vary greatly across different epitopes, yet remain largely conserved across individuals. The emergence of sensitive major histocompatibility complex (MHC) multimer staining protocols has further permitted the accurate measurement of specific T cell populations in the naive pool. Theoretical and experimental studies have indicated that the ability of the T cell repertoire to recognize any novel antigen is essentially determined by the frequency of antigen-specific clonotypes prior to immune challenge. However, it is now possible to extract potentially more useful information from these rich datasets by stratifying for antigen specificity, as exemplified recently in the settings of cytomegalovirus (CMV) infection and ankylosing spondylitis. Previous large-scale studies of immune repertoire structure in health and disease have been limited in the main to analyses of basic parameters, such as repertoire diversity and somatic rearrangement patterns incorporating variable (V), diversity (D), and joining (J) segments of the TCR. The availability of huge volumes of repertoire sequencing (RepSeq) data and a growing curated list of T cell receptor (TCR) sequences with known antigen specificities have enabled quantitative exploration of the adaptive immune system. This inference leads to a new definition of epitope immunogenicity based on specific TCR frequencies, which can be estimated with a high degree of accuracy in silico, thereby providing a novel framework to integrate computational and experimental genomics with basic and translational research efforts in the field of T cell immunology. Our results suggest that the population frequencies of specific T cells are strikingly non-uniform across epitopes that are known to elicit immune responses. We also identified a quantitative link between specific T cell frequencies and the immunogenicity of cognate epitopes presented by defined HLA class I molecules.
The occurrence of certain rearrangements was influenced by ancestry and HLA class I restriction, and umbilical cord blood samples contained higher frequencies of common pathogen-specific TCRs. Our findings revealed a high degree of variability in the prevalence of T cells specific for different antigens that could be explained by the physicochemical properties of the corresponding HLA class I-bound peptides. We also interrogated a database of immunogenic and non-immunogenic peptides is used to link baseline T-cell frequencies with epitope immunogenicity.

We verified our estimates using a publicly available collection of TCR repertoires from healthy individuals. We used a database of TCR sequences with known antigen specificities and a probabilistic TCR rearrangement model to estimate the baseline frequencies of TCRs specific to distinct antigens epitopespecificT-cells. Using recent advances in immune repertoire sequencing technologies, models of the immune receptor rearrangement process, and a database of annotated T cell receptor (TCR) sequences with known specificities, we explored the baseline frequencies of T cells specific for defined human leukocyte antigen (HLA) class I-restricted epitopes in healthy individuals. Adaptive immune responses to newly encountered pathogens depend on the mobilization of antigen-specific clonotypes from a vastly diverse pool of naive T cells.


 0 kommentar(er)
0 kommentar(er)
